A Suite of Ontologies
for Preserving Workflow Research Objects
This web page describes the Research Object and all the resources associated to the paper with the same title currently submitted at the Journal of Web Semantics. The paper describes four ontologies for representing workflows in Research Objects, and includes examples and motivation scenarios.
Abstract
Scientific workflows have, in recent years, gained momentum in modern sciences as one of the popular means for specifying and automating data-driven in silico experiments. The value of scientific workflows also lies in their potential reusability. A workflow provides an explicit specification of an experiment. Once shared, workflow specifications become useful building blocks to be combined or modified for developing new experiments. However, storing specifications is not sufficient for preserving workflows. Scientists need to have an understanding of the workflow and how it may be used and repurposed for their needs. For this purpose, additional resources such as annotations describing the workflow, datasets used and produced by the workflow, and provenance traces recording workflow executions, may be needed to preserve workflows. In this article, we present a suite of ontologies that can be used for preserving scientific workflows. The design of the ontologies was guided by requirements that were raised by scientists involved in the wf4ever project, as well as by our experience in dealing with workflow decay in previous work. The ontologies developed make use of and extend existing well known ontologies, namely the Object Reuse and Exchange (ORE) vocabulary, the Annotation Ontology (AO) and the W3C PROV ontology (PROVO). We illustrate how the ontologies can be utilized using a real-world scenario, in which scientists created a Workflow Research Object for an investigation on the Huntington's disease. We also present the tools we developed for managing Workflow Research Objects.
Inputs and examples of the analysis
Motivation scenario: the Huntington's Disease
Huntington's disease (HD) is the most common inherited neurodegenerative disorder in Europe, affecting 1 out of 10000 people. Although the genetic mutation that causes HD was identified 20 years ago, the downstream molecular mechanisms leading to the HD phenotype are still poorly understood. Relating alterations in gene expression to epigenetic information might shed light on the disease aetiology. In the context of the Wf4ever project, a team of scientists from the Leiden University Medical Centre (HG-LUMC) investigates this hypothesis using workflows.
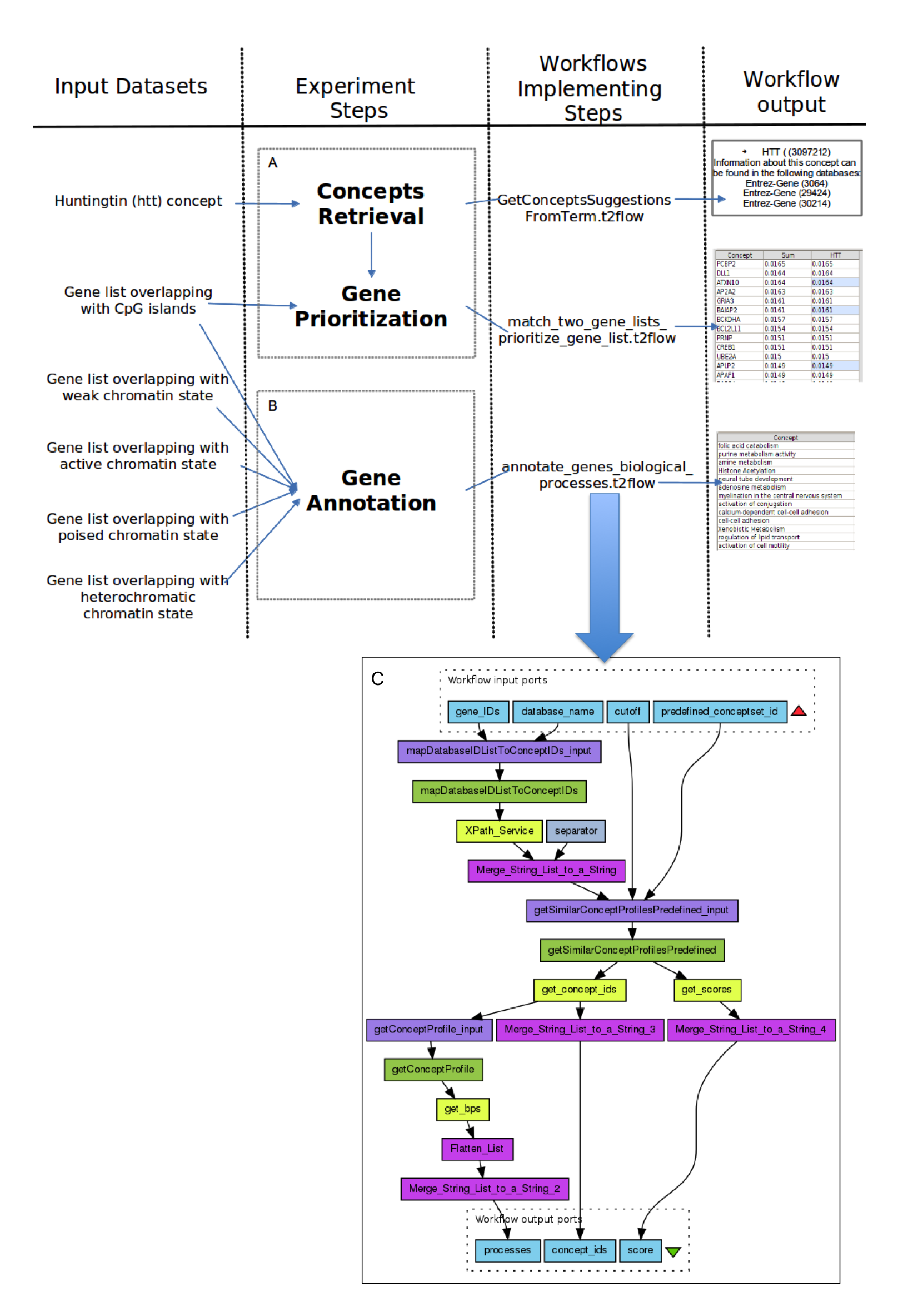
Reused Workflows and Research Objects Figure 1 illustrates the main steps that the bioinformaticians followed in their in silico analysis. The figure sketches out the datasets used as inputs in the analysis, the steps that compose the analysis, and the workflows implementing the steps of the analysis. The bioinformaticians implemented two types of workflows to serve their in silico analysis. The first two workflows, getConceptsSuggestionsFromTerm.t2.flow and match_two_gene_lists_prioritize_gene_list.t2flow are used for gene prioritization, and are executed in sequence.
The third workflow is used to annotate the genes, that are retrieved from five datasets. Once the workflows have been executed, the bioinformaticians analyzed the results to reach their conclusions. The analysis of the workflow results is not illustrated in the sketch depicted in Figure 1 for simplicity.
A research object with executions of the third workflow can be accessed here
Paper results and resources
Our results can be summarized as four main ontologies
- Wfdesc, ontoloy used for specifying workflow descriptions. Here is an example modeling the third workflow of the Research Object.
- Wfprov, ontology used to describe workflow executions. Here is an example modeling an execution of the third workflow included in the Research Object.
- WRO, ontology used to describe general Research Objects and their resources.Here is an example.
- ROEvo, ontology for describing the evolution of Research Objects.Here is an example.
About the authors
| Khalid Belhajjame | Lecturer at the University Paris-Dauphine, where he is a member of the LAMSADE research lab. His research interests lie in the areas of information and knowledge management. In particular, he has made key contributions to the areas of pay-as-you data integration, e-Science, scientific workflow management, provenance tracking and exploitation, and semantic web services. He has published over 50 papers in the aforementioned topics. Most of his research proposals were validated against real-world applications from the fields of astronomy, biodiversity and life sciences. He has participated in multiple European-, French- and UK-funded projects, and have been an active member of theW3C Provenance working group, the NSF funded DataONE working group on scientific workflows and provenance, and more recently the Research Object for Scholarly Communication Community Group. He is also co-leading the provenance benchmarking activity ProvBench, which seeks to produce a family of benchmarks for testing provenance proposals. |
| Jun Zhao | Lecturer at the School of Computing and Communication (Lancaster University), interested in developing novel computing technologies and applications to enable more productive and rigorous scientific research that can make a difference to our lives and well-being. More specifically, her research expertise is in the management and reasoning over provenance information, which records where things come from and how, in order to verify and assess the quality of scientific information resources. |
| Daniel Garijo | PhD student in the Ontology Engineering Group at the Artificial Intelligence Department of the Computer Science Faculty of Universidad Politécnica de Madrid. His research activities focus on e-Science and the Semantic web, specifically on how to increase the understandability of scientific workflows using provenance, metadata, intermediate results and Linked Data |
| Kristina Hettne | Bioinformatician working in the field of biosemantics, passionate about interpreting complex biological data. Through her experience within the Workflow4Ever project, Krisitna has become a strong believer of the necessity of preserving computational experiments for replication, reuse and re-purposing. |
| Raul Palma | Leader of the R&D activities around applied Semantic Web technologies and the participation of the Poznan Supercomputing and Networking Center (PSNC) in different EU projects around these topics, including the preparation of project proposals. As part of this work, I am involved in the specification of software architectures, systems design and API definitions, in addition of leading a team of software engineers and developers. I also collaborate occasionally with the Poznan University of Technology (PUT), where I have participated in other EU project. |
| Eleni Mina | PhD student at the Leiden University Medical Center. |
| Oscar Corcho | Associate Professor at Departamento de Inteligencia Artificial (Facultad de Informática, Universidad Politécnica de Madrid) , and he belongs to the Ontology Engineering Group. His research activities are focused on Semantic e-Science and Real World Internet, although he also works in the more general areas of Semantic Web and Ontological Engineering. In these areas, he has participated in a number of EU projects (Wf4Ever, PlanetData, SemsorGrid4Env, ADMIRE, OntoGrid, Esperonto, Knowledge Web and OntoWeb), and Spanish Research and Development projects (CENITS mIO!, España Virtual and Buscamedia, myBigData, GeoBuddies), and has also participated in privately-funded projects like ICPS (International Classification of Patient Safety), funded by the World Health Organisation, and HALO, funded by Vulcan Inc. |
| Jose Manuel Gomez-Perez | R & D Director at Intelligent Software Components (iSOCO) S.A, where his work focuses on the development and fulfillment of the company's research agenda in target scientific and technological areas, especially in the Semantic Web. His research aims at supporting users in creating, sharing, and exploiting knowledge and spans across different but complementary areas, including: knowledge acquisition, provenance, knowledge visualization and navigation, and applications to intelligent information access and management. Among other positions, he has previously worked as Sr. Research Fellow at Universidad Politecnica de Madrid and Research Manager at iSOCO, and has consulted for companies like British Telecom. |
| Sean Bechhofer | Lecturer in the Information Management Group within the University of Manchester School of Computer Science. His research interests are centered around the technologies required to implement and deliver the Semantic Web. |
| Graham Klyne | Developer currently working at the Image Bioinformatics Group of the zoology department at Oxford University, applying Semantic Web technologies to the development of a semantically-enhanced image database and publishing system (http://www.bioimage.org), working on Web portal technogoes for virtual research envrionments (VREs) for Oxford University Computing Services (OUCS) Research Technologies Service (RTS), and developing embedded web software for a startup company O2M8. His interests include metadata and knowledge based applications using RDF and other Semantic Web technologies, application messaging and asyncronously coupled network systems |
| Carole Goble | Full professor in the University of Manchester School of Computer Science, where she co-leads the Information Management Group. She has worked closely with life scientists for many years and has an international reputation in the Semantic Web, e-Science and Grid communities. Carole is the Director of the myGrid project, a team that produce and use a suite of tools designed to “help e-Scientists get on with science and get on with scientists” |
Acknowledgements
Insert acknowledgemnts here if appropriate
This page is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 2.0 Generic License.

